PDF Parsing
Table of Contents
1 Preparing our data
1.1 Converting PDFs to images
Not all pdfs need to be sent through OCR to extract the text content. If you can click and drag to highlight text in the pdf, then the tools in this library probably aren’t necessary.
This code calls out to pdfimages from Poppler.
def pdf_to_images(pdf_filepath): """ Turn a pdf into images """ directory, filename = os.path.split(pdf_filepath) with working_dir(directory): image_filenames = pdfimages(pdf_filepath) # Since pdfimages creates a number of files named each for there page number # and doesn't return us the list that it created return [os.path.join(directory, f) for f in image_filenames] def pdfimages(pdf_filepath): """ Uses the `pdfimages` utility from Poppler (https://poppler.freedesktop.org/). Creates images out of each page. Images are prefixed by their name sans extension and suffixed by their page number. """ directory, filename = os.path.split(pdf_filepath) filename_sans_ext = filename.split(".pdf")[0] subprocess.run(["pdfimages", "-png", pdf_filepath, filename.split(".pdf")[0]]) image_filenames = find_matching_files_in_dir(filename_sans_ext, directory) logger.debug("Converted {} into files:\n{}".format(pdf_filepath, "\n".join(image_filenames))) return image_filenames def find_matching_files_in_dir(file_prefix, directory): files = [ filename for filename in os.listdir(directory) if re.match(r"{}.*\.png".format(re.escape(file_prefix)), filename) ] return files
1.2 Detecting image orientation and applying rotation.
Tesseract can detect orientation and we can then use ImageMagick’s mogrify to rotate the image.
Here’s an example of the output we get from orientation detection with Tesseract.
➜ example/ tesseract --psm 0 example-000.png - Page number: 0 Orientation in degrees: 90 Rotate: 270 Orientation confidence: 26.86 Script: Latin Script confidence: 2.44
def preprocess_img(filepath): """ Processing that involves running shell executables, like mogrify to rotate. """ rotate = get_rotate(filepath) logger.debug("Rotating {} by {}.".format(filepath, rotate)) mogrify(filepath, rotate) def get_rotate(image_filepath): output = ( subprocess.check_output(["tesseract", "--psm", "0", image_filepath, "-"]) .decode("utf-8") .split("\n") ) output = next(l for l in output if "Rotate: " in l) output = output.split(": ")[1] return output def mogrify(image_filepath, rotate): subprocess.run(["mogrify", "-rotate", rotate, image_filepath])
2 Detecting tables
This answer from opencv.org was heavily referenced while writing the code around table detection: https://answers.opencv.org/question/63847/how-to-extract-tables-from-an-image/.
It’s much easier to OCR a table when the table is the only thing in the image. This code detects tables in an image and returns a list of images of just the tables, no surrounding text or noise.
The blurring, thresholding, and line detection is used here as well as later on for cell extraction. They are good techniques for cleaning an image up in a way that makes things like shape detection more accurate.
def find_tables(image): <<blur>> <<threshold>> <<lines-of-table>> contours, heirarchy = cv2.findContours( mask, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE, ) MIN_TABLE_AREA = 1e5 contours = [c for c in contours if cv2.contourArea(c) > MIN_TABLE_AREA] perimeter_lengths = [cv2.arcLength(c, True) for c in contours] epsilons = [0.1 * p for p in perimeter_lengths] approx_polys = [cv2.approxPolyDP(c, e, True) for c, e in zip(contours, epsilons)] bounding_rects = [cv2.boundingRect(a) for a in approx_polys] # The link where a lot of this code was borrowed from recommends an # additional step to check the number of "joints" inside this bounding rectangle. # A table should have a lot of intersections. We might have a rectangular image # here though which would only have 4 intersections, 1 at each corner. # Leaving that step as a future TODO if it is ever necessary. images = [image[y:y+h, x:x+w] for x, y, w, h in bounding_rects] return images
import cv2 <<detect-table>> image_filename = "resources/examples/example-page.png" image = cv2.imread(image_filename, cv2.IMREAD_GRAYSCALE) image = find_tables(image)[0] cv2.imwrite("resources/examples/example-table.png", image) "resources/examples/example-table.png"
3 OCR tables
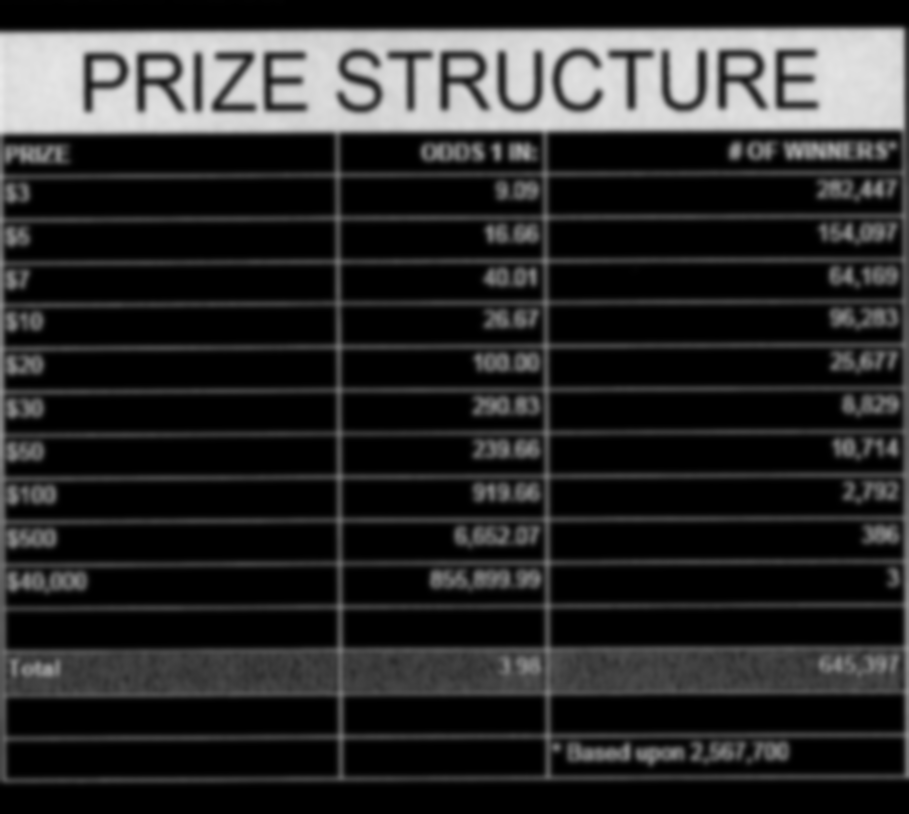
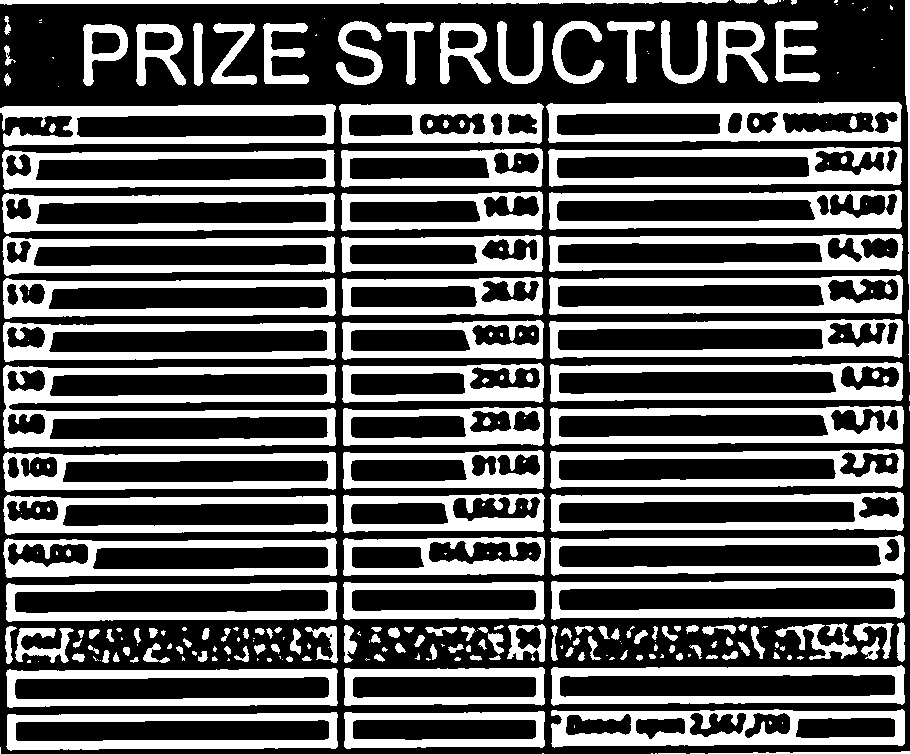
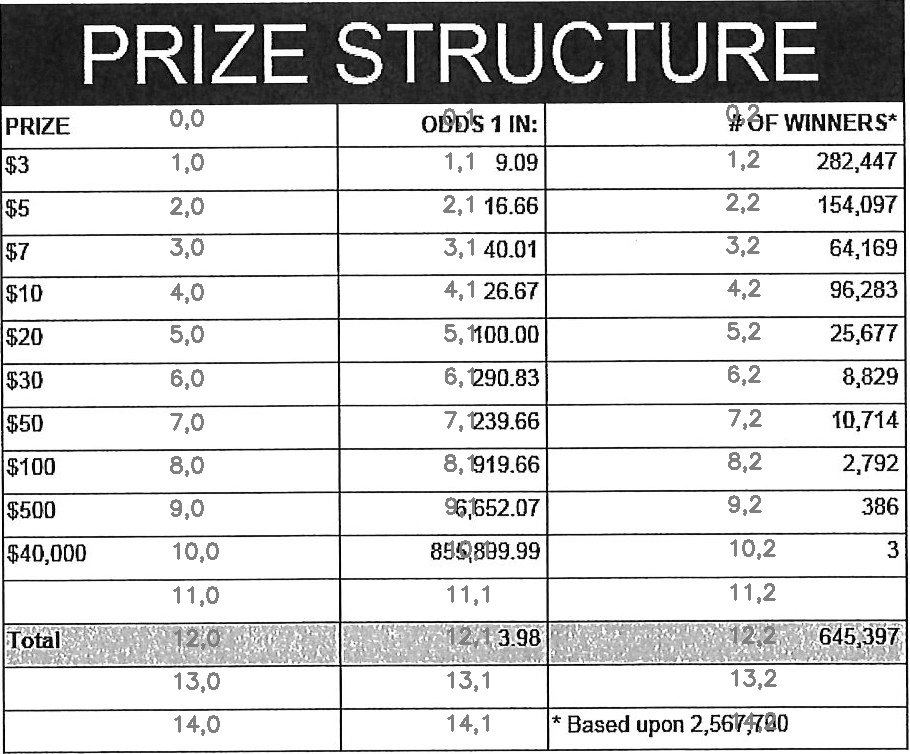
Find the bounding box of each cell in the table. Run tesseract on each cell. Print a comma seperated output.
We’ll start with an image shown at the end of the previous section.
3.0.1 Blur
Blurring helps to make noise less noisy so that the overall structure of an image is more detectable.
That gray row at the bottom is kind of noisy. If we don’t somehow clean it up, OpenCV will detect all sorts of odd shapes in there and it will throw off our cell detection.
Cleanup can be accomplished with a blur followed by some thresholding.
BLUR_KERNEL_SIZE = (17, 17) STD_DEV_X_DIRECTION = 0 STD_DEV_Y_DIRECTION = 0 blurred = cv2.GaussianBlur(image, BLUR_KERNEL_SIZE, STD_DEV_X_DIRECTION, STD_DEV_Y_DIRECTION)
image = ~cv2.imread("resources/examples/example-table.png", cv2.IMREAD_GRAYSCALE) <<blur>> cv2.imwrite("resources/examples/example-table-blurred.png", blurred) "resources/examples/example-table-blurred.png"
3.0.2 Threshold
We’ve got a bunch of pixels that are gray. Thresholding will turn them all either black or white. Having all black or white pixels lets us do morphological transformations like erosion and dilation.
MAX_COLOR_VAL = 255 BLOCK_SIZE = 15 SUBTRACT_FROM_MEAN = -2 img_bin = cv2.adaptiveThreshold( ~blurred, MAX_COLOR_VAL, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY, BLOCK_SIZE, SUBTRACT_FROM_MEAN, )
<<threshold>> cv2.imwrite("resources/examples/example-table-thresholded.png", img_bin) "resources/examples/example-table-thresholded.png"
3.0.3 Finding the vertical and horizontal lines of the table
Note: There’s a wierd issue with the results of the example below when it’s evaluated as part of an export or a full-buffer evaluation. If you evaluate the example by itself, it looks the way it’s intended. If you evaluate it as part of an entire buffer evaluation, it’s distorted.
vertical = horizontal = img_bin.copy() SCALE = 5 image_width, image_height = horizontal.shape horizontal_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (int(image_width / SCALE), 1)) horizontally_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, horizontal_kernel) vertical_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (1, int(image_height / SCALE))) vertically_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, vertical_kernel) horizontally_dilated = cv2.dilate(horizontally_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (40, 1))) vertically_dilated = cv2.dilate(vertically_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (1, 60))) mask = horizontally_dilated + vertically_dilated
<<lines-of-table>> cv2.imwrite("resources/examples/example-table-lines.png", mask) "resources/examples/example-table-lines.png"

3.0.4 Finding the contours
Blurring and thresholding allow us to find the lines. Opening the lines allows us to find the contours.
An “Opening” is an erosion followed by a dilation. Great examples and descriptions of each morphological operation can be found at https://docs.opencv.org/trunk/d9/d61/tutorial_py_morphological_ops.html.
Contours can be explained simply as a curve joining all the continuous points (along the boundary), having same color or intensity. The contours are a useful tool for shape analysis and object detection and recognition.
We can search those contours to find rectangles of certain size.
To do that, we can use OpenCV’s approxPolyEP function. It takes as arguments
the contour (list of contiguous points), and a number representing how different
the polygon perimeter length can be from the true perimeter length of the
contour. 0.1 (10%) seems to be a good value. The difference in perimeter
length between a 4-sided polygon and a 3-sided polygon is greater than 10% and
the difference between a 5+ sided polygon and a 4-sided polygon is less than
10%. So a 4-sided polygon is the polygon with the fewest sides that leaves the
difference in perimeter length within our 10% threshold.
Then we just get the bounding rectangle of that polygon and we have our cells!
We might need to do a little more filtering of those rectangles though. We might have accidentally found some noise such as another image on the page or a title header bar or something. If we know our cells are all within a certain size (by area of pixels) then we can filter out the junk cells by removing cells above/below certain sizes.
contours, heirarchy = cv2.findContours( mask, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE, ) perimeter_lengths = [cv2.arcLength(c, True) for c in contours] epsilons = [0.05 * p for p in perimeter_lengths] approx_polys = [cv2.approxPolyDP(c, e, True) for c, e in zip(contours, epsilons)] # Filter out contours that aren't rectangular. Those that aren't rectangular # are probably noise. approx_rects = [p for p in approx_polys if len(p) == 4] bounding_rects = [cv2.boundingRect(a) for a in approx_polys] # Filter out rectangles that are too narrow or too short. MIN_RECT_WIDTH = 40 MIN_RECT_HEIGHT = 10 bounding_rects = [ r for r in bounding_rects if MIN_RECT_WIDTH < r[2] and MIN_RECT_HEIGHT < r[3] ] # The largest bounding rectangle is assumed to be the entire table. # Remove it from the list. We don't want to accidentally try to OCR # the entire table. largest_rect = max(bounding_rects, key=lambda r: r[2] * r[3]) bounding_rects = [b for b in bounding_rects if b is not largest_rect] cells = [c for c in bounding_rects]
3.0.5 Sorting the bounding rectangles
We want to process these from left-to-right, top-to-bottom.
I’ve thought of a straightforward algorithm for it, but it could probably be made more efficient.
We’ll find the most rectangle with the most top-left corner. Then we’ll find all of the rectangles that have a center that is within the top-y and bottom-y values of that top-left rectangle. Then we’ll sort those rectangles by the x value of their center. We’ll remove those rectangles from the list and repeat.
def cell_in_same_row(c1, c2): c1_center = c1[1] + c1[3] - c1[3] / 2 c2_bottom = c2[1] + c2[3] c2_top = c2[1] return c2_top < c1_center < c2_bottom orig_cells = [c for c in cells] rows = [] while cells: first = cells[0] rest = cells[1:] cells_in_same_row = sorted( [ c for c in rest if cell_in_same_row(c, first) ], key=lambda c: c[0] ) row_cells = sorted([first] + cells_in_same_row, key=lambda c: c[0]) rows.append(row_cells) cells = [ c for c in rest if not cell_in_same_row(c, first) ] # Sort rows by average height of their center. def avg_height_of_center(row): centers = [y + h - h / 2 for x, y, w, h in row] return sum(centers) / len(centers) rows.sort(key=avg_height_of_center)
To test if this code works, let’s try sorting the bounding rectangles and numbering them from right to left, top to bottom.
import cv2 image = cv2.imread("resources/examples/example-table.png", cv2.IMREAD_GRAYSCALE) <<blur>> <<threshold>> <<lines-of-table>> <<bounding-rects>> <<sort-contours>> FONT_SCALE = 0.7 FONT_COLOR = (127, 127, 127) for i, row in enumerate(rows): for j, cell in enumerate(row): x, y, w, h = cell cv2.putText( image, "{},{}".format(i, j), (int(x + w - w / 2), int(y + h - h / 2)), cv2.FONT_HERSHEY_SIMPLEX, FONT_SCALE, FONT_COLOR, 2, ) cv2.imwrite("resources/examples/example-table-cells-numbered.png", image) "resources/examples/example-table-cells-numbered.png"
def extract_cell_images_from_table(image): BLUR_KERNEL_SIZE = (17, 17) STD_DEV_X_DIRECTION = 0 STD_DEV_Y_DIRECTION = 0 blurred = cv2.GaussianBlur(image, BLUR_KERNEL_SIZE, STD_DEV_X_DIRECTION, STD_DEV_Y_DIRECTION) MAX_COLOR_VAL = 255 BLOCK_SIZE = 15 SUBTRACT_FROM_MEAN = -2 img_bin = cv2.adaptiveThreshold( ~blurred, MAX_COLOR_VAL, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY, BLOCK_SIZE, SUBTRACT_FROM_MEAN, ) vertical = horizontal = img_bin.copy() SCALE = 5 image_width, image_height = horizontal.shape horizontal_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (int(image_width / SCALE), 1)) horizontally_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, horizontal_kernel) vertical_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (1, int(image_height / SCALE))) vertically_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, vertical_kernel) horizontally_dilated = cv2.dilate(horizontally_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (40, 1))) vertically_dilated = cv2.dilate(vertically_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (1, 60))) mask = horizontally_dilated + vertically_dilated contours, heirarchy = cv2.findContours( mask, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE, ) perimeter_lengths = [cv2.arcLength(c, True) for c in contours] epsilons = [0.05 * p for p in perimeter_lengths] approx_polys = [cv2.approxPolyDP(c, e, True) for c, e in zip(contours, epsilons)] # Filter out contours that aren't rectangular. Those that aren't rectangular # are probably noise. approx_rects = [p for p in approx_polys if len(p) == 4] bounding_rects = [cv2.boundingRect(a) for a in approx_polys] # Filter out rectangles that are too narrow or too short. MIN_RECT_WIDTH = 40 MIN_RECT_HEIGHT = 10 bounding_rects = [ r for r in bounding_rects if MIN_RECT_WIDTH < r[2] and MIN_RECT_HEIGHT < r[3] ] # The largest bounding rectangle is assumed to be the entire table. # Remove it from the list. We don't want to accidentally try to OCR # the entire table. largest_rect = max(bounding_rects, key=lambda r: r[2] * r[3]) bounding_rects = [b for b in bounding_rects if b is not largest_rect] cells = [c for c in bounding_rects] def cell_in_same_row(c1, c2): c1_center = c1[1] + c1[3] - c1[3] / 2 c2_bottom = c2[1] + c2[3] c2_top = c2[1] return c2_top < c1_center < c2_bottom orig_cells = [c for c in cells] rows = [] while cells: first = cells[0] rest = cells[1:] cells_in_same_row = sorted( [ c for c in rest if cell_in_same_row(c, first) ], key=lambda c: c[0] ) row_cells = sorted([first] + cells_in_same_row, key=lambda c: c[0]) rows.append(row_cells) cells = [ c for c in rest if not cell_in_same_row(c, first) ] # Sort rows by average height of their center. def avg_height_of_center(row): centers = [y + h - h / 2 for x, y, w, h in row] return sum(centers) / len(centers) rows.sort(key=avg_height_of_center) cell_images_rows = [] for row in rows: cell_images_row = [] for x, y, w, h in row: cell_images_row.append(image[y:y+h, x:x+w]) cell_images_rows.append(cell_images_row) return cell_images_rows
<<extract-cells-from-table>> image = cv2.imread("resources/examples/example-table.png", cv2.IMREAD_GRAYSCALE) cell_images_rows = extract_cell_images_from_table(image) cv2.imwrite("resources/examples/example-table-cell-1-1.png", cell_images_rows[1][1]) "resources/examples/example-table-cell-1-1.png"
3.0.6 Cropping each cell to the text
OCR with Tesseract works best when there is about 10 pixels of white border around the text.
Our bounding rectangles may have picked up some stray pixels from the horizontal and vertical lines of the cells in the table. It’s probobly just a few pixels, much fewer than the width of the text. If that’s the case, then we can remove that noise with a simple open morph.
Once the stray border pixels have been removed, we can expand our border using
openMakeBorder.
def crop_to_text(image): kernel = cv2.getStructuringElement(cv2.MORPH_CROSS, (4, 4)) opened = cv2.morphologyEx(~image, cv2.MORPH_OPEN, kernel) contours, hierarchy = cv2.findContours(opened, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE) bounding_rects = [cv2.boundingRect(c) for c in contours] # The largest contour is certainly the text that we're looking for. largest_rect = max(bounding_rects, key=lambda r: r[2] * r[3]) x, y, w, h = largest_rect cropped = image[y:y+h, x:x+w] bordered = cv2.copyMakeBorder(cropped, 5, 5, 5, 5, cv2.BORDER_CONSTANT, None, 255) return bordered
import cv2 <<crop-to-text>> image = cv2.imread("resources/examples/example-table-cell-1-1.png", cv2.IMREAD_GRAYSCALE) image = crop_to_text(image) cv2.imwrite("resources/examples/example-table-cell-1-1-cropped.png", image) "resources/examples/example-table-cell-1-1-cropped.png"

3.0.7 OCR each cell
If we cleaned up the images well enough, we might get some accurate OCR!
There is plenty that could have gone wrong along the way.
The first step to troubleshooting is to view the intermediate images and see if there’s something about your image that is obviously abnormal, like some really thick noise or a wrongly detected table.
If everything looks reasonable but the OCR is doing something like turning a period into a comma, then you might need to do some custom Tesseract training.
def crop_to_text(image): kernel = cv2.getStructuringElement(cv2.MORPH_CROSS, (4, 4)) opened = cv2.morphologyEx(~image, cv2.MORPH_OPEN, kernel) contours, hierarchy = cv2.findContours(opened, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE) bounding_rects = [cv2.boundingRect(c) for c in contours] # The largest contour is certainly the text that we're looking for. largest_rect = max(bounding_rects, key=lambda r: r[2] * r[3]) x, y, w, h = largest_rect cropped = image[y:y+h, x:x+w] bordered = cv2.copyMakeBorder(cropped, 5, 5, 5, 5, cv2.BORDER_CONSTANT, None, 255) return bordered def ocr_image(image, config): cropped = crop_to_text(image) return pytesseract.image_to_string( ~cropped, config=config )
import pytesseract image = cv2.imread("resources/examples/example-table-cell-1-1.png", cv2.IMREAD_GRAYSCALE) <<ocr-image>> ocr_image(image, "--psm 7")
9.09
4 Files
4.1 setup.py
import setuptools with open("README.md", "r") as fh: long_description = fh.read() setuptools.setup( name="example-pkg-YOUR-USERNAME-HERE", # Replace with your own username version="0.0.1", author="Example Author", author_email="author@example.com", description="A small example package", long_description=long_description, long_description_content_type="text/markdown", url="https://github.com/pypa/sampleproject", packages=setuptools.find_packages(), classifiers=[ "Programming Language :: Python :: 3", "License :: OSI Approved :: MIT License", "Operating System :: OS Independent", ], python_requires='>=3.6', )
4.2 table_image_ocr
4.2.1 table_image_ocr/__init__.py
4.2.2 table_image_ocr/util.py
from contextlib import contextmanager import functools import logging import os import tempfile from bs4 import BeautifulSoup as bs import requests logger = get_logger() @contextmanager def working_dir(directory): original_working_dir = os.getcwd() try: os.chdir(directory) yield directory finally: os.chdir(original_working_dir) def download(url, filepath): response = request_get(url) data = response.content with open(filepath, "wb") as f: f.write(data) def make_tempdir(identifier): return tempfile.mkdtemp(prefix="{}_".format(identifier))
4.2.3 table_image_ocr/prepare_pdfs.py
Takes a variable number of pdf files and creates images out of each page of the
file using pdfimages from Poppler. Images are created in the same directory
that contains the pdf.
Prints each pdf followed by the images extracted from that pdf followed by a blank line.
python -m pdf.prepare_pdfs /tmp/file1/file1.pdf /tmp/file2/file2.pdf ...
import argparse import logging import os import re import subprocess import sys from pdf.util import request_get, working_dir, download, make_tempdir logger = get_logger() parser = argparse.ArgumentParser() parser.add_argument("files", nargs="+") def main(files): pdf_images = [] for f in files: pdf_images.append((f, pdf_to_images(f))) for pdf, images in pdf_images: for image in images: preprocess_img(image) for pdf, images in pdf_images: print("{}\n{}\n".format(pdf, "\n".join(images))) def pdf_to_images(pdf_filepath): """ Turn a pdf into images """ directory, filename = os.path.split(pdf_filepath) with working_dir(directory): image_filenames = pdfimages(pdf_filepath) # Since pdfimages creates a number of files named each for there page number # and doesn't return us the list that it created return [os.path.join(directory, f) for f in image_filenames] def pdfimages(pdf_filepath): """ Uses the `pdfimages` utility from Poppler (https://poppler.freedesktop.org/). Creates images out of each page. Images are prefixed by their name sans extension and suffixed by their page number. """ directory, filename = os.path.split(pdf_filepath) filename_sans_ext = filename.split(".pdf")[0] subprocess.run(["pdfimages", "-png", pdf_filepath, filename.split(".pdf")[0]]) image_filenames = find_matching_files_in_dir(filename_sans_ext, directory) logger.debug("Converted {} into files:\n{}".format(pdf_filepath, "\n".join(image_filenames))) return image_filenames def find_matching_files_in_dir(file_prefix, directory): files = [ filename for filename in os.listdir(directory) if re.match(r"{}.*\.png".format(re.escape(file_prefix)), filename) ] return files def preprocess_img(filepath): """ Processing that involves running shell executables, like mogrify to rotate. """ rotate = get_rotate(filepath) logger.debug("Rotating {} by {}.".format(filepath, rotate)) mogrify(filepath, rotate) def get_rotate(image_filepath): output = ( subprocess.check_output(["tesseract", "--psm", "0", image_filepath, "-"]) .decode("utf-8") .split("\n") ) output = next(l for l in output if "Rotate: " in l) output = output.split(": ")[1] return output def mogrify(image_filepath, rotate): subprocess.run(["mogrify", "-rotate", rotate, image_filepath]) if __name__ == "__main__": args = parser.parse_args() main(args.files)
4.2.4 table_image_ocr/extract_tables.py
. ~/.virtualenvs/lotto_odds/bin/activate
python -m pdf.extract_tables "resources/examples/example-page.png"
import argparse import os import cv2 parser = argparse.ArgumentParser() parser.add_argument("files", nargs="+") def main(files): results = [] for f in files: directory, filename = os.path.split(f) image = cv2.imread(f, cv2.IMREAD_GRAYSCALE) tables = find_tables(image) files = [] for i, table in enumerate(tables): filename_sans_extension = os.path.splitext(filename)[0] table_filename = "{}-table-{:03d}.png".format(filename_sans_extension, i) table_filepath = os.path.join(directory, table_filename) files.append(table_filepath) cv2.imwrite(table_filepath, table) results.append((f, files)) for image_filename, table_filenames in results: print("{}\n{}\n".format(image_filename, "\n".join(table_filenames))) def find_tables(image): BLUR_KERNEL_SIZE = (17, 17) STD_DEV_X_DIRECTION = 0 STD_DEV_Y_DIRECTION = 0 blurred = cv2.GaussianBlur(image, BLUR_KERNEL_SIZE, STD_DEV_X_DIRECTION, STD_DEV_Y_DIRECTION) MAX_COLOR_VAL = 255 BLOCK_SIZE = 15 SUBTRACT_FROM_MEAN = -2 img_bin = cv2.adaptiveThreshold( ~blurred, MAX_COLOR_VAL, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY, BLOCK_SIZE, SUBTRACT_FROM_MEAN, ) vertical = horizontal = img_bin.copy() SCALE = 5 image_width, image_height = horizontal.shape horizontal_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (int(image_width / SCALE), 1)) horizontally_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, horizontal_kernel) vertical_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (1, int(image_height / SCALE))) vertically_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, vertical_kernel) horizontally_dilated = cv2.dilate(horizontally_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (40, 1))) vertically_dilated = cv2.dilate(vertically_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (1, 60))) mask = horizontally_dilated + vertically_dilated contours, heirarchy = cv2.findContours( mask, cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE, ) MIN_TABLE_AREA = 1e5 contours = [c for c in contours if cv2.contourArea(c) > MIN_TABLE_AREA] perimeter_lengths = [cv2.arcLength(c, True) for c in contours] epsilons = [0.1 * p for p in perimeter_lengths] approx_polys = [cv2.approxPolyDP(c, e, True) for c, e in zip(contours, epsilons)] bounding_rects = [cv2.boundingRect(a) for a in approx_polys] # The link where a lot of this code was borrowed from recommends an # additional step to check the number of "joints" inside this bounding rectangle. # A table should have a lot of intersections. We might have a rectangular image # here though which would only have 4 intersections, 1 at each corner. # Leaving that step as a future TODO if it is ever necessary. images = [image[y:y+h, x:x+w] for x, y, w, h in bounding_rects] return images if __name__ == "__main__": args = parser.parse_args() files = args.files main(files)
4.2.5 table_image_ocr/extract_cells_from_table.py
. ~/.virtualenvs/lotto_odds/bin/activate
python -m pdf.extract_cells_from_table "resources/examples/example-table.png"
import os import sys import cv2 import pytesseract def main(f): results = [] directory, filename = os.path.split(f) table = cv2.imread(f, cv2.IMREAD_GRAYSCALE) rows = extract_cell_images_from_table(table) cell_img_dir = os.path.join(directory, "cells") os.makedirs(cell_img_dir, exist_ok=True) for i, row in enumerate(rows): for j, cell in enumerate(row): cell_filename = "{:03d}-{:03d}.png".format(i, j) path = os.path.join(cell_img_dir, cell_filename) cv2.imwrite(path, cell) print(cell_filename) def extract_cell_images_from_table(image): BLUR_KERNEL_SIZE = (17, 17) STD_DEV_X_DIRECTION = 0 STD_DEV_Y_DIRECTION = 0 blurred = cv2.GaussianBlur(image, BLUR_KERNEL_SIZE, STD_DEV_X_DIRECTION, STD_DEV_Y_DIRECTION) MAX_COLOR_VAL = 255 BLOCK_SIZE = 15 SUBTRACT_FROM_MEAN = -2 img_bin = cv2.adaptiveThreshold( ~blurred, MAX_COLOR_VAL, cv2.ADAPTIVE_THRESH_MEAN_C, cv2.THRESH_BINARY, BLOCK_SIZE, SUBTRACT_FROM_MEAN, ) vertical = horizontal = img_bin.copy() SCALE = 5 image_width, image_height = horizontal.shape horizontal_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (int(image_width / SCALE), 1)) horizontally_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, horizontal_kernel) vertical_kernel = cv2.getStructuringElement(cv2.MORPH_RECT, (1, int(image_height / SCALE))) vertically_opened = cv2.morphologyEx(img_bin, cv2.MORPH_OPEN, vertical_kernel) horizontally_dilated = cv2.dilate(horizontally_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (40, 1))) vertically_dilated = cv2.dilate(vertically_opened, cv2.getStructuringElement(cv2.MORPH_RECT, (1, 60))) mask = horizontally_dilated + vertically_dilated contours, heirarchy = cv2.findContours( mask, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE, ) perimeter_lengths = [cv2.arcLength(c, True) for c in contours] epsilons = [0.05 * p for p in perimeter_lengths] approx_polys = [cv2.approxPolyDP(c, e, True) for c, e in zip(contours, epsilons)] # Filter out contours that aren't rectangular. Those that aren't rectangular # are probably noise. approx_rects = [p for p in approx_polys if len(p) == 4] bounding_rects = [cv2.boundingRect(a) for a in approx_polys] # Filter out rectangles that are too narrow or too short. MIN_RECT_WIDTH = 40 MIN_RECT_HEIGHT = 10 bounding_rects = [ r for r in bounding_rects if MIN_RECT_WIDTH < r[2] and MIN_RECT_HEIGHT < r[3] ] # The largest bounding rectangle is assumed to be the entire table. # Remove it from the list. We don't want to accidentally try to OCR # the entire table. largest_rect = max(bounding_rects, key=lambda r: r[2] * r[3]) bounding_rects = [b for b in bounding_rects if b is not largest_rect] cells = [c for c in bounding_rects] def cell_in_same_row(c1, c2): c1_center = c1[1] + c1[3] - c1[3] / 2 c2_bottom = c2[1] + c2[3] c2_top = c2[1] return c2_top < c1_center < c2_bottom orig_cells = [c for c in cells] rows = [] while cells: first = cells[0] rest = cells[1:] cells_in_same_row = sorted( [ c for c in rest if cell_in_same_row(c, first) ], key=lambda c: c[0] ) row_cells = sorted([first] + cells_in_same_row, key=lambda c: c[0]) rows.append(row_cells) cells = [ c for c in rest if not cell_in_same_row(c, first) ] # Sort rows by average height of their center. def avg_height_of_center(row): centers = [y + h - h / 2 for x, y, w, h in row] return sum(centers) / len(centers) rows.sort(key=avg_height_of_center) cell_images_rows = [] for row in rows: cell_images_row = [] for x, y, w, h in row: cell_images_row.append(image[y:y+h, x:x+w]) cell_images_rows.append(cell_images_row) return cell_images_rows if __name__ == "__main__": main(sys.argv[1])
5 Utils
The following code lets us specify a size for images when they are exported to html.
Org supports specifying an export size for an image by putting the #+ATTR_HTML:
:width 100px before the image. But since our images are in a results drawer, we
need a way for our results drawer to do that for us automatically.
Adding #+ATTR_HTML after the beginning of the result block introduces a new
problem. Org-babel no longer recognizes the result as a result block and doesn’t
remove it when a src block is re-evaluated, so we end up just appending new
results on each evaluation.
There is nothing configurable that will tell org-babel to remove our line. But
we can define a function to do some cleanup and then add it as a before hook
with advice-add.
(concat "#+ATTR_HTML: :width " width " :height " height "\n[[file:" text "]]")
(defun remove-attributes-from-src-block-result (&rest args) (let ((location (org-babel-where-is-src-block-result)) (attr-regexp "[ ]*#\\+ATTR.*$")) (when location (save-excursion (goto-char location) (when (looking-at (concat org-babel-result-regexp ".*$")) (next-line) (while (looking-at attr-regexp) (kill-whole-line))))))) (advice-add 'org-babel-remove-result :before #'remove-attributes-from-src-block-result) (advice-add 'org-babel-execute-src-block :before #'remove-attributes-from-src-block-result)